Research
Characterizing Spermatogenesis through the Inactivation of Uba2 in the SUMOylation Cycle and Its Meiotic Arrest
By: Hanna Wiesen, Margarita Vigodner
Abstract
Purpose: To investigate the alterations in the expression of noncoding, micro, and small RNA expression during treatment with oncolytic reovirus in KRAS-mutated colorectal cancer. Methods: Oncolytic reovirus treatment was administered in phase 1 clinical trial (NCT01274624) for 5 days every 28 days, and blood samples were collected before the administration of the reovirus and 48 h, 8 days, and 15 days after its administration on day 1. Data from the blood samples were sorted using Transcriptome Analysis Software (TAC) 4.0, where a two-tailed t-test and a fold change filter were used to ascertain which sample signals had a statistically significant relative fold change of greater than 2 at multiple timepoints before or after oncolytic reovirus administration. Results: The long noncoding RNA’s RP11-332M2.1 (−6.1 x), LINC01506 (−16.18 x), and LINC00534 (−1.94 x) were downregulated at 48 h after reovirus administration [p < 0.05]. ncRNA’s EPB41L4A-AS1 (−6.34 x, 48 h; 11.99 x, day 8), JAK2 (2.2 x, 48 h; −2.23 x, day 8), ANXA4 (20.47 x, day 8; −7.54 x, day 15), and PCDH9 (−2.09, day 8; 1.82 x, day 15) were affected by the reovirus treatment and reflected the progress of the treatment [p < 0.05]. The small RNA SNORA26 (−1.59 x, day 8) was downregulated 48 h after the reovirus administration [p < 0.05]. The microRNA MIR-4461 (6.18 x, day 8; −3.76 x, day 15) was also affected by the reovirus administration [p < 0.05]. Conclusion: The administration of oncolytic reovirus to treat KRAS-mutated colorectal cancer is reflected in a noncoding RNA profile, and expression levels of the ncRNAs in that profile may thus be able to be used as a potential predictive marker for reovirus-treated colorectal cancer.

Figure 1. Stages of Meiosis I [1]
In prophase I, the nuclear envelope is broken down and the chromosome condenses and takes up 90% of the process of meiosis [2]. Prophase I has its own five stages: leptotene, zygotene, pachytene, diplotene, and diakinesis (Figure 2).

Figure 2. Stages of Meiotic Prophase I [3]
Leptotene is when individual chromosomes begin to condense inside of the nucleus. The homologous chromosomes align during zygotene, forming a tetrad of all four sister chromatids. Additionally, the synaptonemal complex forms between the homologous chromosomes, linking the chromosomes together tightly by various proteins such as SCP3, synaptonemal protein 3 [2]. This complex is composed of two side-by-side protein axes with a recombination nodule in the middle. This nodule catalyzes crossing over, the random exchange of genetic information between two homologous chromosomes. Crossing over occurs in pachytene, the next stage. In diplotene, the following stage, the synaptonemal complex is broken down and the regions of DNA where crossing over occurred are identified as the chiasmata, held together by proteins [2]. Additionally, the DNA uncoils a little, allowing for minor quantities of RNA transcription (Figure 3) [4].
Diakinesis is the final stage of prophase I, where the chromosomes recondense. Prophase I is the longest stage of meiosis. Cells that have completed prophase I have reached mid-late meiosis because the rest of meiosis I and II occur quickly [2].
Meiosis I continues with metaphase I, where the homologous chromosomes lineup in a random order in the middle of the cell, at the metaphase plate. In anaphase I, the homologous chromosomes are pulled apart by spindle fibers, followed by telophase I, which completes the split of the original germ cell.
Meiosis II parallels meiosis I, with two major exceptions. Crossing over does not occur, and instead of pairs of homologous chromosomes being separated, the sister chromatids are pulled apart additionally. Meiosis II contains the same stages as meiosis I [6].
Meiosis of male gametes is the first step in sperm production, which takes place in the testes, an organ which plays a significant role in male reproduction. Each testis is around 3cm wide and 5cm long [7]. It is made up of three parts: the tunica vaginalis, the serous covering of the testes, the tunica albuginea, the fibrous covering of the testes, and the septa, tissue barriers that extend from the tunica albuginea (Figure 4) [8].
Within the testes are around 250 testicular lobules, which contain 1-4 tightly coiled seminiferous tubules; when extended the total tubule length is between 30-91 centimeters [9]. The seminiferous tubules produce and facilitate sperm cell maturation, as well as secrete hormones.
Within the testes, each tubule contains Leydig, myoid, and Sertoli cells, three somatic cells which are regulated by hormones secreted by different regions of the hypothalamus. Gonadotropin releasing hormone, GNRH, is secreted from the hypothalamus, and stimulates the anterior pituitary gland to secrete luteinizing hormone, LH, and follicle stimulating hormone, FSH. LH and FSH are released into the bloodstream and act as part of the endocrine system by acting upon testicular somatic cells [10]. LH stimulates Leydig cells, which are found in the space between the tubules, adjunct to the blood vessels. The Leydig cells synthesize and secrete testosterone [11]. Testosterone is concentrated around the tubules to support gamete maturation and circulates the bloodstream to support secondary male characteristics. When testosterone levels within the blood become too high, this hormone acts as negative feedback to the hypothalamus and anterior pituitary to limit secretion of GNRH and LH. FSH acts on the Sertoli cells, which regulates sperm development and synthesizes three separate hormones [10]. The first hormone produced is an androgen-binding hormone, which is crucial to the progression of gametes through spermatogenesis. The second hormone is estrogen. The third hormone secreted is inhibin, which inhibits the production of FSH in the anterior pituitary gland, also acting as a negative feedback mechanism (Figure 5) [10, 12].
The second main role of the testes is to facilitate an environment in which sperm cells can mature into spermatids which can fertilize an egg. Gamete maturation occurs inside of the seminiferous tubules and is regulated by Sertoli cells [13]. As the gametes mature, they move from the outer areas of the tubules to the inner areas between the Sertoli cells. Sertoli cells secrete crucial nutrients for sperm cell development, creating a “nursery” in the tubules (Figure 6) [14].
Sperm cells mature through a process called spermatogenesis. Before a boy begins puberty, all his sperm cells are type A spermatogonia; these cells have spherical, oval nuclei, and are located by the basement membrane, alongside the Sertoli cells. Once puberty begins, the type A spermatogonia acts as stem cells, dividing asymmetrically into type A spermatogonia and type B spermatogonia [16]. Type B spermatogonia, also known as primary spermatocytes, are the cells that enter meiosis, beginning spermatogenesis (Figure 7) [17].
The primary spermatocytes progress through meiosis, producing round spermatids at the end of meiosis II; these cells have moved towards the lumen of the seminiferous tubules (Figure 8) [16].
The round spermatids then begin spermiogenesis, the process through which round spermatids mature into elongated spermatids ready to fertilize an egg cell [16, 20]. The first stage of spermiogenesis is the golgi phase, when the acrosomal vesicle forms and the centrioles move towards the opposite end of the cell. Step two is the acrosomal stage, when the nucleus is elongated and condensed, the centrioles form a flagellum, and the mitochondria surround the flagella’s base. The third stage is the maturation phase, where the spermatozoa’s cell body is shed, and the spermatid is released into the lumen of the tubules as fully mature sperm (Figure 9) [21].
The sperm’s structure consists of three main parts: the head, midpiece, and tail. The head contains the nucleus and the acrosome, encompassing enzymes necessary for fertilization. The midpiece of the sperm has mitochondria located by the base of its tail, which provides energy for flagellar movement. The tail contains the flagella which propels the sperm up through the female reproductive tract (Figure 10) [18].
Each step of spermatogenesis and spermiogenesis is crucial for functional, mature sperm. Additionally, the crosstalk between cells within the tubules is crucial for sperm development. Any error in any of the stages or hormone pathways mentioned above can lead to infertility, which is why a strong understanding of the tubules, and the breakdown of spermatogenesis is crucial [23].
Infertility
Infertility is when a couple has difficulty conceiving after 12 months of trying without intervention [24]. Infertility can occur when a couple is trying for their first child or any subsequent child, and affects 8-12% of couples [25]. There is a 30% chance that there are issues within the woman’s reproductive system, a 30% chance there are issues within the man’s reproductive system, and a 30% chance there are issues in both man and woman or that the infertility is idiopathic, meaning that the cause is unknown (Figure 11) [25].
Male factors contributes to one third of infertility cases and impact about 50% of all infertility cases. Some of these factors include poor sperm motility, count, and density, hormonal disorders, and testicular trauma, among others. Couples may also have prior existing medical conditions that may limit the man’s ability to produce viable sperm [26]. As a result of modern medicine, there are many treatments that have helped couples combat infertility. However, there are still 33% of couples who are struggling with idiopathic infertility [27]. This statistic emphasizes the importance of studying spermatogenesis and other potential factors causing male infertility.
Post Translational Modifications
Spermatogenesis is tightly regulated by post translational modifications (PTMs), the addition of proteins or radical groups to other proteins post translation. Without PTMs, many proteins are not functional; because of this, the addition or removal of such PTMs regulate protein function. Some examples of PTMs include phosphorylation, methylation, ubiquitination, and SUMOylation (Figure 12).
There have been many studies that have examined the effects of phosphorylation and ubiquitination regulation on spermatogenesis [29, 30]. However, until recently, the relationship between SUMOylation and spermatogenesis has not been researched.
SUMOylation is the cycle which regulates DNA transcription through the addition or removal of SUMO peptides to various proteins. The SUMOylation cycle consists of three steps; the first step is activation, where SUMO binds to Uba2 activating enzyme, the first enzyme. This allows SUMO to bind to the second enzyme, Ubc9, in the conjugation stage. Ubc9 brings together SUMO peptide sequence with its substrate, the third enzyme, through ligation. The SUMOylation cycle is also reversible through the usage of SUMO-specific isopeptidases; these enzymes remove SUMO from the substrate, regulating SUMO’s cellular activity (Figure 13) [31].
This process occurs throughout the body but occurs significantly more frequently within the testes; however, its importance and role in spermatogenesis and infertility has not yet been fully characterized [33].
Previous Laboratory Data
Previously, Dr. Vigodner’s lab has conducted extensive research on the relationship between SUMOylation’s role in testicular cells and spermatogenesis. Dr. Vigodner and her colleagues conducted research in vitro, in vivo, and ex vivo. In vitro research used cell lines that mimic primary cells only to a certain degree. Ex vivo research involves experiments in the dish using primary spermatocytes purified from the mouse testes. In vivo research involved transgenic mice.
This research focused on studying SUMOylation in different cell types. While studying meiosis, the group showed that when ginkgolic acid, GA, a compound that inhibits SUMOylation, was added to mouse spermatocytes, the GA prevented the spermatocytes from having chromatin condensation, disassembly of the synaptonemal complex, and the completion of meiosis [34, 35]. These results suggested that SUMOylation was required for the completion of meiosis in spermatocytes. To clarify its role, the next step was to study SUMOylation in vivo and determine how SUMOylation inhibition affects the mice’s fertility.
DNA Excision through Cre-LoxP System
Dr. Vigodner analyzed the effects of inhibiting UBA-2 function within the SUMOylation cycle through transgenic mice. In vivo research that manipulates DNA and alters gene and protein expression in mice produces transgenic mice [36]. Since the SUMOylation cycle is crucial to produce viable offspring, there was no effective way to study this process in adult testes. Therefore, to study the effects of SUMOylation on male mice meiosis, the Cre-LoxP system was used to genetically inactivate SUMO-activating enzyme Uba2 specifically in spermatocytes.
The Cre-LoxP System effectively inactivates the transcription of a specific protein in vivo. A mouse is bred with LoxP sites of DNA inserted before and after the gene of interest [gene X]. Cre, an enzyme that is encoded by a separate gene, recognized these LoxP sites. The Cre enzyme acts as scissors and removes gene X from between the LoxP sites on the genome, effectively removing gene X in that mouse (Figure 14) [36].
There are various Cre enzymes for different tissues. For example, to biologically cut, or excise, a gene in liver cells, a mouse with the liver Cre promoter would be bred with a mouse whose gene of interest is bound by LoxP. The offspring would be a heterozygous Cre-LoxP mouse, containing Cre and its promoter, one regular gene, and one gene contains LoxP; therefore, this mouse would have one gene with only the LoxP sites and a second gene with the gene of interest (Figure 15) [36].
This heterozygous Cre-LoxP mouse would breed with a mouse that is homozygous for LoxP genes, meaning that both of its alleles for the gene of interest are in between LoxP sites. 25% of this breeding’s offspring will be homozygous for the LoxP sites on the gene and hemizygous for Cre and its promoter; this mouse is homozygous for the excision in the specific tissue (Figure 16) [36].
To study the role of SUMOylation in meiosis in vivo, Dr. Vigodner bred Cre-LoxP mice where the LoxP sites were placed on either side of exon 3 for the protein Uba2, the activation enzyme in step 1 of the SUMOylation cycle. Stra-8, the promoter for Cre that is expressed only in meiotic cells, activates Cre to excise exon 3 from the mice’s DNA (Figure 17) [19].
As a result, Uba2 was not properly transcribed and was therefore nonfunctional in the experimental mice. Given that Uba2 is the first step in the SUMOylation cycle, and the SUMOylation process plays a critical role in meiotic cells, the deletion of exon 3 through the Cre-LoxP breeding method using Stra-8 inhibited the SUMOylation in male meiotic cells (Figure 18) [19].
Morphological Analysis of Transgenic Mice
When the experimental mice were produced, their fertility, body, and testicular weight were evaluated. The transgenic mice were infertile and did not produce pups after 6 weeks, while the control mice produced multiple litters. The body weight of both the control and experimental mice were around the same (Table 1).
However, there was a significant different between the experiment and control mice’s testicular weight. The weight of the experimental mice’s testes were much lower than those of the control mice (Table 2).
Additionally, when the testes from both mice were examined side by side, it was clear that the testes of the experimental mice were significantly smaller than those of the control mice.
Current Goals
The infertility and significantly decreased testicular weight in the transgenic mice suggests that spermatogenesis was arrested. Therefore, the goals of this research were to characterize spermatogenesis in transgenic mice to gain a better understanding of causes for infertility, and to characterize spermatogenesis in transgenic mice in comparison to the control mice.
Methods and Materials
Cross sections of the control and experimental mice’s testes were examined under a microscope. Spermatocytes, round spermatids, and elongated spermatids were counted. Immunohistochemistry was used to identify the presence of H1T in transgenic cells; H1T is a marker of mid-late meiosis [37, 38]. A series of deparaffinization, rehydration steps, and staining were used to identify the presence of H1T; immunohistochemistry is further explained in previously published articles [39]. H1T is a marker of slightly unraveled DNA, occurring during diplotene of prophase I [40].
Western Blots were used by running proteins on a gel electrophoresis, transferring them to a membrane, and identifying the proteins through antibodies [34]. Anti-SUMO antibody, Synaptonemal Complex Protein 3, and tubulin were run on a Western Blot. The Western Blot technique is discussed in previously published articles [41].
RNA Scope and BaseScope Assay Technique
RNA Scope Assay, developed in 2011, is a technique using in situ hybridization to recognize target mRNA sequences [42]. “Z” probes are used to locate the target mRNA. The Z probe is composed of RNA bases, recognizes complimentary RNA sequences with over 300 bases, and has 3 unique areas [42]. The first area is at the “bottom” of the Z and contains between 8-25 bases, called the target binding site. The target binding site is complementary to the target RNA sequence. The middle of the Z serves as a “linker” between the target binding site and the pre-amplifier binding site. The pre-amplifier binding site is the top of the “Z” and has about 14 base pairs [43]. When 2 specific Z probes are hybridized together and bind to the RNA target sequence, they form a base region with about 50 bases complementary to the target RNA and a 28 base pre-amplifier tail. To distinguish between target and nontarget RNA, the Z’s must be hybridized together to provide a full pre-amplifier binding site; nontarget RNA can bind to a single Z probe containing 14 bases, half of the required bases to complete a full pre-amplifier binding site. Because of this, only Z pairs can produce signal amplification [43].
RNA Scope requires at least 7 Z pairs to produce a signal but may need up to 20 Z pairs to successfully produce a signal [42]. To produce a signal amplification, a Z pair must hybridize and bind to their target RNA. The amplifier binds to its complete binding site of 28 bases at the top of the Z pairs, containing multiple binding sites for the ISH. To recognize the presence of the target RNA sequence, fluorescent or chromogenic probes bind to the amplifier sites to produce a visible result. Each RNA sequence can be identified by a visible by using a standard microscope (Figure 18) [43].
RNA BaseScope, developed in 2016, is an advanced form of RNA Scope that allowed for the detection of smaller RNA targets. Both RNA assays utilize Z pairs, amplifiers, and ISH [42]. However, in contrast to RNA Scope Assays, BaseScope Assays can detect RNA sequences containing 50-300 base pairs with only 1-3 Z pairs, as opposed to 7. Additionally, while RNA Scope can detect strands of mRNA, BaseScope can detect splice variants and individual point mutations within an mRNA sequence [45]. Splice variants are changes in DNA that result in a change of exon and intron splicing within the RNA sequence (Figure 19) [46].
In this research, RNA BaseScope Assays were used to detect splice variants of exon 3 in Uba2 proteins. The BaseScope assays used chromogenic mRNA probes of exon 3 of Uba2 to identify the presence of RNA splice variant deletion of exon 3 in Uba2 in experimental and control mice as a reflection of functional Uba2.
Results
Morphological and Quantitative Analysis of Seminiferous Tubules in Transgenic Mice
The cross section of the testes in the control and experimental mice were visually analyzed through a counting of spermatocytes, round spermatids, and elongated spermatids. The different types of cells were clear in the microscopic images taken of each cross section. In the control mice, the tubules are large, they are full of cells, and all types of sperm cells are easily visible. These tubules have spermatogonia, spermatocytes, round spermatids, and elongated spermatids in the lumen of the tubules. In the experimental mice, however, the tubules clearly illustrate that the cells have arrested spermatogenesis. The tubules are significantly smaller and have degenerated. Additionally, there are very few round or elongated spermatids within the tests. These results suggest arrested spermatogenesis (Figure 20).
Indeed, quantitative analysis of the two cross sections of the tubules supported these results. This analysis showed a decrease in spermatocytes (Table 3a), a drastic decrease in round spermatids (Table 3b), and almost no elongated spermatids (Table 3c). Because there were so few round and elongated spermatids, these results indicate that the meiotic arrest occurred at the spermatocyte stage.
Immunohistochemistry Analysis of Spermatocytes
Since spermatogenesis is arrested during meiosis at the spermatocyte stage, we wondered whether the spermatocytes present in the transgenic tubules were early or late spermatocytes. We used immunohistochemistry (IHC) to identify the presence of H1T within the transgenic mice presented positive results. H1T is a marker of mid-late meiosis [37]; the presence of H1T on the slides is indicated by a brown staining of the nuclei of spermatocytes. Most of the spermatocytes indicated positive H1T results (Figure 21).

Figure 3. Synaptonemal Complex [5]
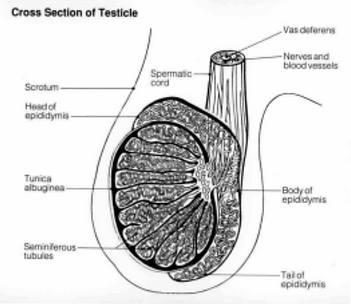
Figure 4. Cross Section of the Testicle [1]
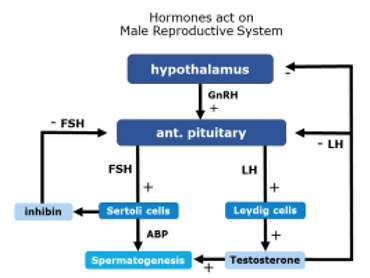
Figure 5. Hormonal Regulation in Male Reproduction [12]
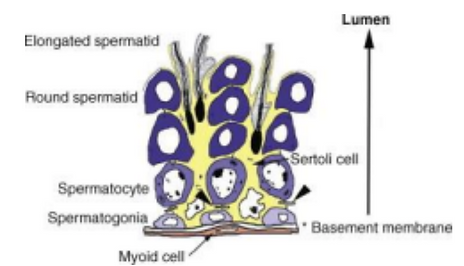
Figure 6. Testicular Cells within the Seminiferous Tubule [15]

Figure 7. Spermatogenesis Diagram. Germ cells develop from Type A spermatogonia to Type B spermatogonia, also called primary spermatocytes. Primary spermatocytes enter meiosis I, which produces secondary spermatocytes. Secondary spermatocytes enter meiosis II, which produces round spermatids. Round spermatids mature into spermatozoa through spermiogenesis [18].
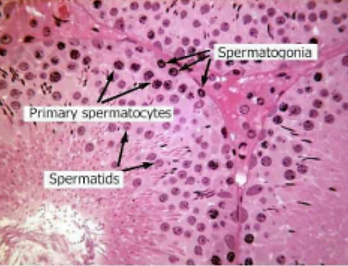
Figure 8. Cross Section of Healthy, Fertile Seminiferous Tubules. Spermatogonia are found by the basal membrane of the tubules. As they mature into spermatocytes and then into spermatids, the cells are moved into the center of the tubules by Sertoli cells [19].
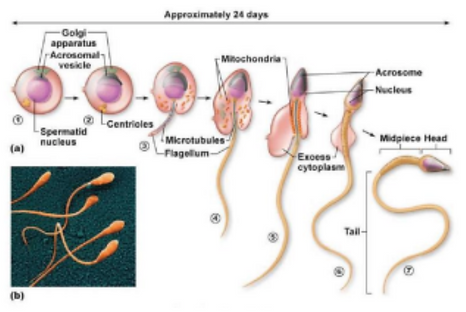
Figure 9. Spermiogenesis Diagram. Round spermatids develop into elongated spermatids and then spermatozoa [22].
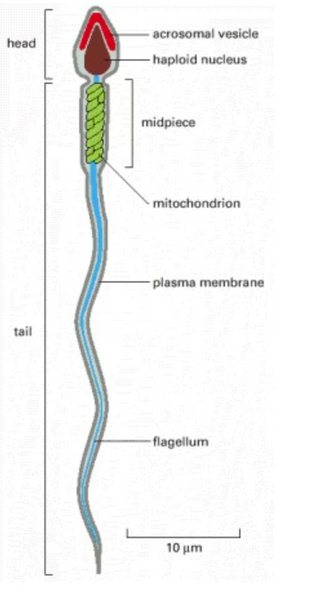
Figure 10. Sperm Cell Anatomy [18]

Figure 11. Breakdown of Factors for Infertility [26]
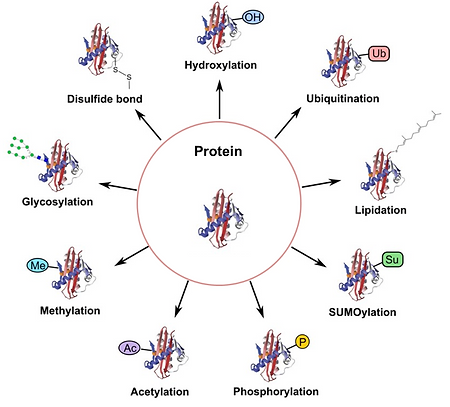
Figure 12. Diagram of Post Translation Modifications [28]
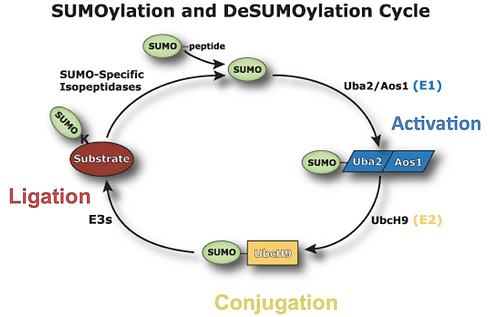
Figure 13. The SUMOylation Cycle [32]
Figure 14. Cre-LoxP Excising a Gene [36]

Figure 15. Diagram of the Production of Cre-LoxP Transgenic Mouse [36]
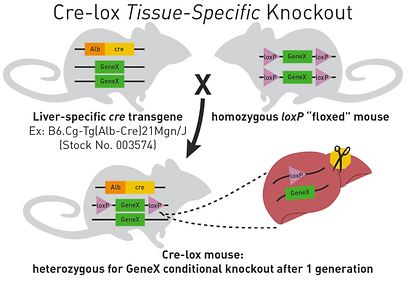
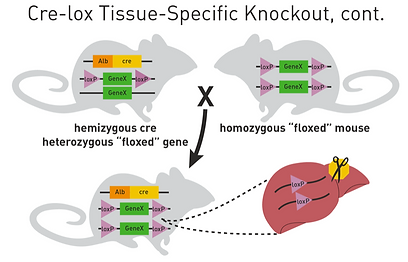
Figure 16. Diagram of the Production of Transgenic Experimental Mice with Cre gene and LoxP sites of both ends of the gene. [36]
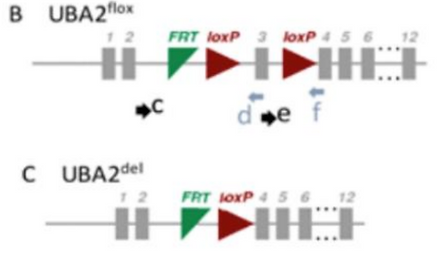
Figure 17. Diagram of Uba2 Gene with LoxP sites on both sides of Exon 3

Figure 18. Double Z Probe used in RNA Scope Assays. The hybridization of the Z pair forms the pre-amplifier binding site, which allows the for the binding of the amplifier. The fluorescent or chromogenic probes provide visual signal amplification [44].
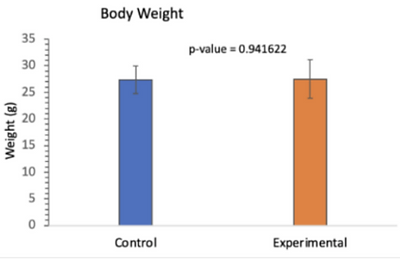
Table 1. Table Comparing the Body Weight of Control and Experiment Mice. There was no significant difference between the body weight of the control and experimental mice [19].
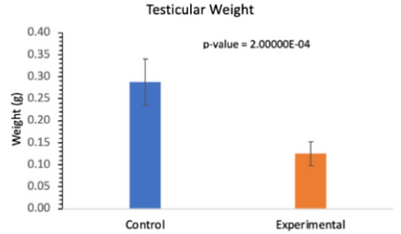
Table 2. Table Comparing the Testicular Weight of Control and Experimental Mice. There was a significant difference between the testicular weight of the control and experimental mice [19].
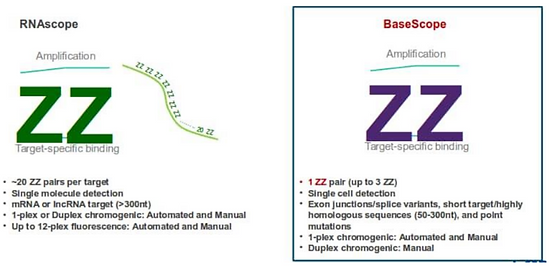
Figure 19. Comparison Between RNA Scope and RNA BaseScope. RNA BaseScope uses the same techniques as RNA Scope but requires fewer Z pairs and can detect shorter and more specific RNA sequences [47].

Figure 20. Cross Section of the Seminiferous Tubules in Control and Experimental Mice. The tubules in the experimental mice are smaller and contain fewer gamete cells than the control tubules.

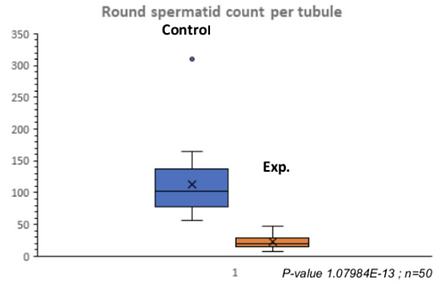
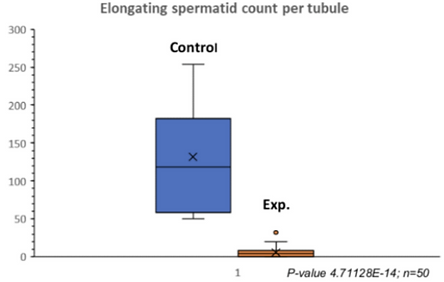
Table 3a. Quantitative Analysis of Spermatocytes in Control and Experimental Tubules. There was a significant decrease in spermatocytes in the experimental tubules.
Table 3b. Quantitative Analysis of Round Spermatids in Experimental and Control Tubules. There was a drastic decrease in round spermatids in the experimental tubules.
Table 3c. Quantitative Analysis of Elongated Spermatids in Experimental and Control Tubules. There were almost no elongated spermatids in the experimental tubules.
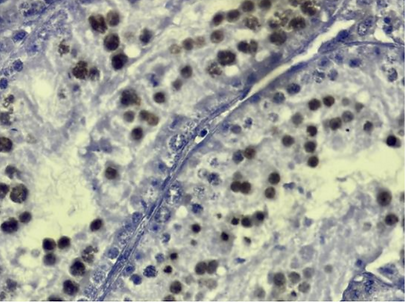
Figure 21. H1T Analysis of Transgenic Tubules. IHC was used to identify the presence of H1T in spermatocytes, a marker of mid-late meiosis. Most of the spermatocytes indicated positive results for H1T staining.
Assessment of Efficiency of Uba2 Inactivation
To assess the efficiency of Uba2 inactivation, we used a Uba2 antibody to analyze if Uba2 was still expressed in the spermatocytes of transgenic mice. We had hoped not to see the presence of Uba2 proteins in transgenic mice since they were the target of inactivation. The presence of brown spermatocytes indicates a positive Uba2 antibody result. The control cells showed positive Uba2 results (Figure 22a). Although we assumed that there would not be any expression of Uba2 in the transgenic mice, there were significant amounts of Uba2 positive spermatocytes as well as Uba2 negative spermatocytes (Figure 22b). Preliminary statistical analysis suggested that only around 50% of transgenic spermatocytes had successful excision of exon 3 of Uba2 and presented a negative Uba2 result.
To clarify whether the excision of exon 3 effectively inhibited Uba2 translation, the RNA BaseScope Assay was used on testicular tissue sections of the control and experimental mice to detect splice variants. Only some control cells indicated the presence of exon 3 of Uba2, when we assumed that all the control cells would have exon 3. Additionally, even though we did not expect to see any RNA probe in the experimental cells like the Uba2 antibody, some of the transgenic cells also showed the presence of exon 3 of Uba2. Red dots on the images indicate a Uba2 positive result (Figure 23).
SUMOylation Analysis
Since the transgenic mice were supposed to lack SUMOylation activating enzyme, Uba2, we hoped to see the defect in the SUMOylation cycle (Figure 24).
The Western Blot was composed of a mixed germ cell fraction which had been separated from Sertoli cells by differential plating from 4 control mice and 4 experimental mice. The proteins with a higher molecular weight are at the top of the blot, while the proteins with a lower molecular weight are at the bottom. A SUMO antibody was used on the membrane to identify protein-bound SUMO and free SUMO. Almost all the control cells had SUMO-bound proteins and little free SUMO, while the transgenic cells had a significant increase in free SUMO and decrease of protein-bound SUMO. Synaptonemal Complex Protein 3 (SCP3) was also used on the blot as a marker of spermatocytes. These results indicate that there were more SCP3 proteins found within the tests of the transgenic mice. Additionally, tubulin antibody was used as a loading control (Figure 25).
Discussion
Characterization of spermatogenesis
The analysis of the different sperm cells through statistical analysis indicates that meiosis was inhibited due to the lack of round and elongated spermatids. There is current analysis of spermatogonia and the preliminary data which may suggest that there are no changes in spermatogonia. Based on these finding, we suggested that the arrest in spermatogenesis happened during meiosis.
To better understand at what stage of meiosis the inactivation of SUMOylation and meiotic arrest occurs, the H1T marker was used to try and locate the point of inactivation through IHC. H1T was specifically used because it is a marker of mid-late meiosis. Most of the spermatocytes in the transgenic mice were stained brown, highlighting the presence of H1T and the cell’s progression until mid-late meiosis. Based on the data collected and from preliminary statistics, this confirmed that spermatocytes begin to enter mid-late meiosis before their meiotic arrest.
Confirming Inactivation of SUMOylation as a Result of the Cre-LoxP System
The Uba2 antibody was used to confirm that the meiotic arrest was due to the excision of exon 3 of Uba2 and as a result, degradation of Uba2 mRNA and the Uba2 protein. Our original assumption was that the control mice would have a completely positive result for Uba2 while the transgenic mice would have a completely negative result for Uba2. Although the control mice indicated the expected result, the transgenic mice showed the presence of Uba2 in many of the spermatocytes. The cells that were negative for Uba2 antibody reflected a successful Cre-LoxP excision of exon 3. Despite around 50% of the cells in the transgenic mice containing exon 3 and producing functional Uba2, spermatogenesis was inhibited and the mice were infertile. This reflects the importance of the hormonal feedback mechanisms and crosstalk between germ and testicular cells described above. Since SUMOylation was inhibited in many germ cells, the crosstalk with Sertoli cells was most likely affected. Therefore, Sertoli cells were not producing enough growth factors. Overall, the results demonstrated that the Cre-LoxP system was not 100% successful and suggested that when a large germ cell fraction is nonfunctional, the entire tubule’s metabolism is affected.
The goal for using RNA BaseScope was to confirm the excision of exon 3 from Uba2, indicating the success of the Cre-LoxP system. We had hoped to observe mRNA in the control spermatocytes and a lack of mRNA in the transgenic mice due to the lack of Uba2. However, mRNA for Uba2 were identified within some of the transgenic cells. Additionally, many of the control cells were negative for the mRNA probe. The varied results within the transgenic cells supported the Uba2 antibody data, proving that the Cre-LoxP was not 100% successful. However, it is important to note that mRNA is not a stable molecule. The preparation for tissue fixation, among other steps, significantly affects mRNA stability. As a result, the control cells were not 100% positive for the mRNA exon 3 probe. Although it seemed that the transgenic mice had fewer positive spermatocytes, the quantification of the results is difficult with such heterogeneity of the staining.
Confirmation SUMOylation Inhibition
The Western Blot was used to confirm the inhibition of SUMOylation in the germ cell fraction of the transgenic mice. The anti-SUMO antibody recognized free SUMO and SUMO that was bound to other proteins of different molecular weight. The results highlighted the significant decrease in SUMOylated isoform and an increase in the amount of free SUMO within all 4 mice, as compared to the control mice. Since Uba2 was nonfunctional, the SUMOylation cycle was significantly inhibited, thereby arresting meiosis.
SCP3 was used as another marker of spermatocytes. SCP3 is present in cells during prophase I, when the synaptonemal complex is forming and is functioning. Since SCP3 is only present in spermatocytes, we had assumed that there would be less SCP3 in the transgenic mice due to the meiotic inhibition; however, our results showed more SCP3 in the experimental cells than the control cells. We then realized that the fraction of germ cells between the control and experimental samples was varied. The control sample contained cells at various maturation points, so its germ cell fraction was composed of spermatocytes, round, and elongated spermatids. The experimental sample, on the other hand, contained mostly spermatocytes, with negligible amounts of spermatids. Its germ cell fraction contained many more spermatocytes than its control counterparts. SCP3 confirmed that the arrest took place during mid-late meiosis and that the experimental samples contained almost no spermatids. Anti-tubulin antibody was also run on the membrane as a loading control since it detects all proteins on the membrane.
Conclusion
In conclusion, the cell specific inactivation of Uba2 using Stra-8 promoter resulted in arrested spermatogenesis in mid-late meiosis and inhibited SUMOylation within male germ cells, causing complete infertility in male mice. Further research should uncover the downstream targets that were affected by the inhibited SUMOylation and the meiotic arrest. Additionally, biopsies from infertile patients with arrested spermatogenesis (including those arrested at the spermatocyte stage) can be screened for mutations in Uba2 and other genes related to SUMOylation.
Acknowledgments
Dr. Cynthia Wachtell
Thank you for your dedication to the S. Daniel Abraham Honors Program at Stern College for Women. Taking part in this program has allowed me to reach my full potential by providing me countless opportunities to broaden my education. I am proud and grateful to have been part of this program.
Dr. Margarita Vigodner
Thank you for your guidance throughout my time at Stern College. I am beyond grateful for your support and your dedication to my success as a student; your continued sensitivity and understanding has been immeasurable. Thank you for providing the opportunity for me to take part in the groundbreaking research in your lab and cultivate my skills in the laboratory setting. Additionally, thank you for your mentorship throughout the process of writing my thesis.
My Family
Thank you for your boundless love and support throughout my college career. I am grateful for your continuous encouragement throughout the process of writing my honors thesis.
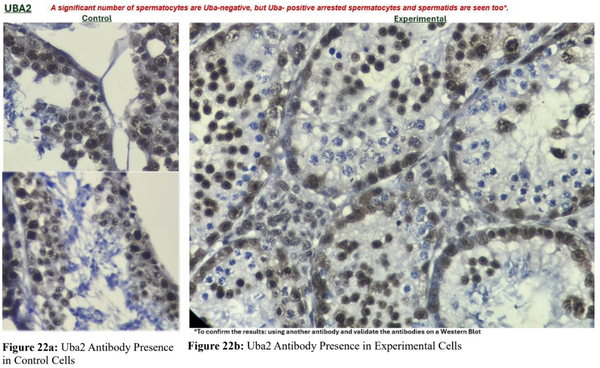
Figure 22. Spermatocyte Analysis for Uba2 using Uba2 Antibody. (22a) The control cells were positive for Uba2. (22b) Only around 50% of experimental cells were negative for Uba2.
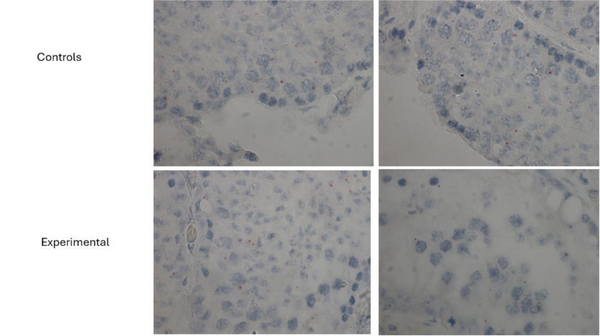
Figure 23. RNA BaseScope Analysis of Exon 3 Probe in Control and Experimental Cells. The chromogenic probes used indicated the presence functional Uba2 in control and experimental cells.

Figure 24. Target of Uba2 Inactivation within the SUMOylation Cycle [32]

Figure 25. Western Blot Analysis of SUMOylated Protein, Free SUMO, SCP3, and Tubulin in Control and Experimental Cells. The control cells had significantly more SUMO-bound proteins and less free SUMO, while the experimental cells had no SUMO-bound proteins and significantly more free SUMO. The experimental cells had more SCP3 than the control cells. Tubulin was used as a marker for equal loading.
References
1.Singh, Natasha Ramroop. “1.3 Meiosis.” Introduction to Genetics, Thompson Rivers University, 1 Feb. 2023,
opengenetics.pressbooks.tru.ca/chapter/meiosis-i/.
2. Alberts, Bruce. “Meiosis.” Molecular Biology of the Cell. 4th Edition., U.S. National Library of Medicine, 1 Jan.
1970, www.ncbi.nlm.nih.gov/books/NBK26840/.
3. “Stages of Meiotic Prophase I: BioRender Science Templates.” Stages of Meiotic Prophase I | BioRender Science
Templates, www.biorender.com/template/stages-of-meiotic-prophase-i.
4. Hill, M.A. “Cell Division - Meiosis.” Embryology, 2 Mar. 2024,
embryology.med.unsw.edu.au/embryology/index.php?title=Cell_Division_-+_Meiosis#Diplotene.
5. Hill, M.A. “1.3: Meiosis.” Biology LibreTexts, Libretexts, 2 Mar. 2024,
bio.libretexts.org/Bookshelves/Genetics/Introduction_to_Genetics_%28Singh%29 /01%3A_Mendels_First_Law_and_Meiosis/1.03%3A_Meiosis.
6. Chapter 9: Concept 9.6, bodell.mtchs.org/OnlineBio/BIOCD/text/chapter9/concept9.6.html.
7. “Testes.” Testes | SEER Training, NIH National Cancer Institute,
training.seer.cancer.gov/anatomy/reproductive/male/testes.html.
8.Tiwana, Manpreet S. “Anatomy, Abdomen and Pelvis: Testes.” StatPearls , U.S. National Library of Medicine, 15
Apr. 2023, www.ncbi.nlm.nih.gov/books/NBK470201/figure/article-33979.image.f2/.
9. Countouris, N., and A.F. Holstein. “Wieviel lobuli testis enthält Ein Menschlicher Hoden? Nachuntersuchung eines
Alten problems.” Andrologia, vol. 17, no. 6, 24 Apr. 2009, pp. 525–531, https://doi.org/10.1111/j.1439-0272.1985.tb01707.x.
10. Gurung, Purnima. “Physiology, Male Reproductive System.” StatPearls, U.S. National Library of Medicine, 1 May
2023, www.ncbi.nlm.nih.gov/books/NBK538429/.
11. Zhou, Rui, et al. “The roles and mechanisms of leydig cells and myoid cells in regulating spermatogenesis.”
Cellular and Molecular Life Sciences, vol. 76, no. 14, 12 Apr. 2019, pp. 2681–2695, https://doi.org/10.1007/s00018-019-03101-9.
12. Bastawros, Hala. “Hormonal Regulation in the Male Reproduction.” Human Reproduction A Clinical Approach,
Iowa State University Digital Press, 12 Dec. 2023, iastate.pressbooks.pub/humanreproduction/chapter/new/.
13. Smith, Lee B, and William H Walker. “The regulation of spermatogenesis by androgens.” Seminars in cell &
developmental biology vol. 30 (2014): 2-13. doi:10.1016/j.semcdb.2014.02.012
14. Wong, Wah J. “Histology, Sertoli Cell.” StatPearls, U.S. National Library of Medicine, 14 Nov. 2022,
www.ncbi.nlm.nih.gov/books/NBK560631/.
15. Yoshida, Shosei. “Heterogeneous, dynamic, and stochastic nature of mammalian spermatogenic stem cells.”
Current Topics in Developmental Biology, 2019, pp. 245–285, https://doi.org/10.1016/bs.ctdb.2019.04.008.
16. Suede, Samah H. “Histology, Spermatogenesis.” StatPearls , U.S. National Library of Medicine, 6 Mar. 2023,
www.ncbi.nlm.nih.gov/books/NBK553142/.
17. Gilbert, Scott F. “Spermatogenesis.” Developmental Biology. 6th Edition., U.S. National Library of Medicine, 1
Jan. 1970, www.ncbi.nlm.nih.gov/books/NBK10095/.
18. Alberts, Bruce. “Sperm.” Molecular Biology of the Cell. 4th Edition., U.S. National Library of Medicine, 1 Jan.
1970, www.ncbi.nlm.nih.gov/books/NBK26914/.
19. Previous Lab Data
20. O’Donnell, Liza. “Mechanisms of spermiogenesis and spermiation and how they are disturbed.” Spermatogenesis,
vol. 4, no. 2, 4 Mar. 2014, https://doi.org/10.4161/21565562.2014.979623.
21. Koita, Irfana. “Spermiogenesis: What Is It, Symptoms and Treatment.” Top Doctors, Top Doctors
https://www.topdoctors.co.uk, 6 May 2023, www.topdoctors.co.uk/medical-dictionary/spermiogenesis#.
22. “Spermatogenesis.” Spermatogenesis - an Overview | ScienceDirect Topics,
www.sciencedirect.com/topics/agricultural-and-biological-sciences/spermatogenesis.
23. Tüttelmann, Frank, et al. “Disorders of spermatogenesis.” Medizinische Genetik, vol. 30, no. 1, 26 Feb. 2018, pp.
12–20, https://doi.org/10.1007/s11825-018-0181-7.
24. “Infertility.” World Health Organization, World Health Organization,
www.who.int/news-room/fact-sheets/detail/infertility.
25. Ombelet, Willem. “WHO fact sheet on infertility gives hope to millions of infertile couples worldwide.” Facts,
views & vision in ObGyn vol. 12,4 249-251. 8 Jan. 2020
26. “How Common Is Infertility?” CNY Fertility, 12 July 2024, www.cnyfertility.com/how-common-is-infertility/.
27. “Infertility: Causes & Treatment.” Cleveland Clinic, 13 June 2024,
my.clevelandclinic.org/health/diseases/16083-infertility.
28. “Strategies for Post-Translational Modifications (PTMS).” Creative Proteomics Blog, 4 June 2018,
www.creative-proteomics.com/blog/index.php/strategies-for-post-translational%20modifications-ptms/.
29. Xiong, Yi, et al. “Ubiquitin-proteasome system–regulated protein degradation in spermatogenesis.” Cells, vol. 11,
no. 6, 21 Mar. 2022, p. 1058, https://doi.org/10.3390/cells11061058.
30. Zhang, Xuedi, et al. “Broad phosphorylation mediated by testis-specific serine/threonine kinases contributes to
spermiogenesis and male fertility.” Nature Communications, vol. 14, no. 1, 6 May 2023, https://doi.org/10.1038/s41467-023-38357-0.
31. Han, Zhi-Jian et al. “The post-translational modification, SUMOylation, and cancer (Review).” International
journal of oncology vol. 52,4 (2018): 1081-1094. doi:10.3892/ijo.2018.4280
32. Scandurra, Amy, and Hai Nguyen. SUMOylaLon Status and Effects of SUMOylaLon on DAX-1, USFCA,
repository.usfca.edu/cgi/viewcontent.cgi?article=1219&context=artsci_stu.
33. Vigodner, Margarita et al. “Identification of sumoylated targets in proliferating mouse spermatogonia and human
testicular seminomas.” Asian journal of andrology vol. 22,6 (2020): 569-577. doi:10.4103/aja.aja_11_20
34. Applebaum, Noa et al. “Phosphoproteome analysis of the crosstalk between sumoylation and phosphorylation in
mouse spermatocytes.” Biochemical and biophysical research communications vol. 681 (2023): 194-199. doi:10.1016/j.bbrc.2023.09.029
35. National Research Council. “Genetically Altered Mice: A Revolutionary Research Resource.” Sharing Laboratory
Resources: Genetically Altered Mice: Summary of a Workshop Held at the National Academy of Sciences, March 23-24, 1993., U.S. National Library of Medicine, 1 Jan. 1994, www.ncbi.nlm.nih.gov/books/NBK231336/.
36. Kelmenson, Peter. “Cre Lox Breeding for Beginners, Part 1.” The Jackson Laboratory, 23 Sept. 2011,
www.jax.org/news-and-insights/jax-blog/2011/september/cre-lox-breeding.
37. Wolfe, Steven A., and Sidney R. Grimes. “Histone H1T: A tissue‐specific model used to study transcriptional
control and nuclear function during cellular differentiation.” Journal of Cellular Biochemistry, vol. 53, no. 2, Oct. 1993, pp. 156–160, https://doi.org/10.1002/jcb.240530208.
38. Magaki, Shino et al. “An Introduction to the Performance of Immunohistochemistry.” Methods in molecular
biology (Clifton, N.J.) vol. 1897 (2019): 289-298. doi:10.1007/978-1-4939-8935-5_25
39.Schafler, Eric D., et al. “UXT is required for spermatogenesis in mice.” PLOS ONE, vol. 13, no. 4, 12 Apr. 2018,
https://doi.org/10.1371/journal.pone.0195747.
40. Mahadevan, Iyer Aditya, et al. “Linker Histone Variant H1T Is Closely Associated with Repressed Repeat-Element
Chromatin Domains in Pachytene Spermatocytes - Epigenetics & Chromatin.” BioMed Central, BioMed Central, 4 Mar. 2020, epigeneticsandchromatin.biomedcentral.com/articles/10.1186/s13072-020-00335-x.
41.Yang, Ping-Chang, and Tahrin Mahmood. “Western blot: Technique, theory, and trouble shooting.” North American
Journal of Medical Sciences, vol. 4, no. 9, 2012, p. 429, https://doi.org/10.4103/1947-2714.100998.
42. “RNAscope, BaseScope and miRNAscope Assays.” Advanced Cell Diagnostics,
acdbio.com/rnascope-basescope-and-mirnascope-assays.
43. “ACD RNAscope in Situ Hybridization (ISH) Technology Overview.” YouTube, Advanced Cell Diagnostics, 26
Aug. 2015, www.youtube.com/watch?v=CPgZEf6EyJQ.
44. George, Logan, et al. “Intracellular RNA-tracking methods.” Open Biology, vol. 8, no. 10, Oct. 2018,
https://doi.org/10.1098/rsob.180104.
45. “Basescope Assays.” Advanced Cell Diagnostics, acdbio.com/basescope-assays-0.
46. “NCI Dictionary of Genetics Terms.” Comprehensive Cancer Information - NCI,
www.cancer.gov/publications/dictionaries/genetics-dictionary/def/splice-site variantwgAEAAYgAQyBwgAEAAYgAQyBggBEEUYOTIICAIQABgWGB4yCAgDE AAYFhgeMgoIBBAAGA8YFhgeMgoIBRAAGA8YFhgeMg0IBhAAGIYDGIAEGIoF Mg0IBxAAGIYDGIAEGIoFqAIAsAIA&sourceid=chrome&ie=UTF-8.
47. “Advanced Cell Diagnostics.” Prima Scientific, en.prima-sci.com/17139384/advanced-cell-diagnostics.
